
Tau lesions, characterized by Tau protein aggregation, are a group of heterogeneous neurodegenerative diseases. The most common Tau degenerative diseases include Alzheimer's disease (AD) and subtypes of frontotemporal degenerative disease (frontotemporal lobar degeneration with Tau pathology, referred to as FTLD-Tau) with Tau lesions, such as Pick's disease (PiD), cortical basal degeneration (CBD), progressive supranuclear paralysis (PSP) and so on.
The Tau protein is encoded by a single MAPT gene and selectively spliced by exon 10 to form six isomers, including three (3R) or four (4R) microtubule binding repeats. There are three subtypes of Tau protein disease, namely, 3R, 4R and 3R/4R mixed type, which show different Tau protein fiber structures under frozen electron microscopy. Therefore, the Tau filaments in AD (3R/4R), PiD (3R) and CBD (4R) are also different in structure, among which the cases with familial frontotemporal lobe degeneration are 4R specific. The lack of a good 4R Tau research model is an important reason for limiting the occurrence and development of related diseases and the discovery of treatments.
Neurons derived from human induced pluripotent stem cells (Human induced pluripotent stem cell, referred to as hiPSC) are of little value in simulating nervous system diseases, including but not limited to Tau disease. Combined with CRISPR-Cas9 technology, the neuron platform derived from iPSC can achieve accurate disease modeling and functional genomics analysis through allelic modification, so that disease-related regulatory factors can be identified. However, even after long-term culture, the expression of 4R Tau in iPSC-derived neurons is very low, so it is not suitable to simulate 4R Tau diseases such as PSP. The low level of Tau containing exon 10 also limits their ability to mimic dominant familial FTLD-Tau mutations in exon 10. In addition, it is difficult to reproduce stable Tau aggregation in human iPSC-derived neurons. Although insoluble Tau aggregation was not observed in MAPT-P301L or MAPT-IVS10+16 iPSC neurons, a small number of Tau inclusion bodies could be observed after 120 days. An important factor leading to this phenomenon may be the lack of 4R Tau in neurons derived from iPSC.
Recently, the Shiaoching Gong and Li Gan research groups from Weill Cornell Medicine in New York published an article entitled "Human iPSC 4R tauopathy model uncovers modifiers of tau propagation" on Cell, and successfully established a robust and scalable human iPSC 4R Tau disease model.

The team constructed an engineered human-induced pluripotent stem cell line that expressed 4R Tau and 4R Tau carrying a P301S MAPT mutation. Through the induction of Tau fibril seeds (seeding), the progressive accumulation of Tau inclusion bodies was shown in 4R-P301S neurons, and the phenotypic features of Tau protein disease were replicated, including transcriptome characteristics, autophagy accumulation and reduced neuronal activity.
Through CRISPR interference (CRISPRi) screening of genes related to Tau pathology, more than 500 genetic modifiers inducing Tau transmission were identified, including genes in the cascade of retromer VPS29 and UFMylation. In the brains of patients with progressive supranuclear paralysis (PSP) and Alzheimer's disease (AD), the UFMylation cascade changes in neurons with neurofilament tangles. Finally, the inhibition of UFMylation cascade in vitro and in vivo blocked the transmission of Tau protein. This model provides a powerful platform for finding new treatment strategies for 4R Tau protein disease.
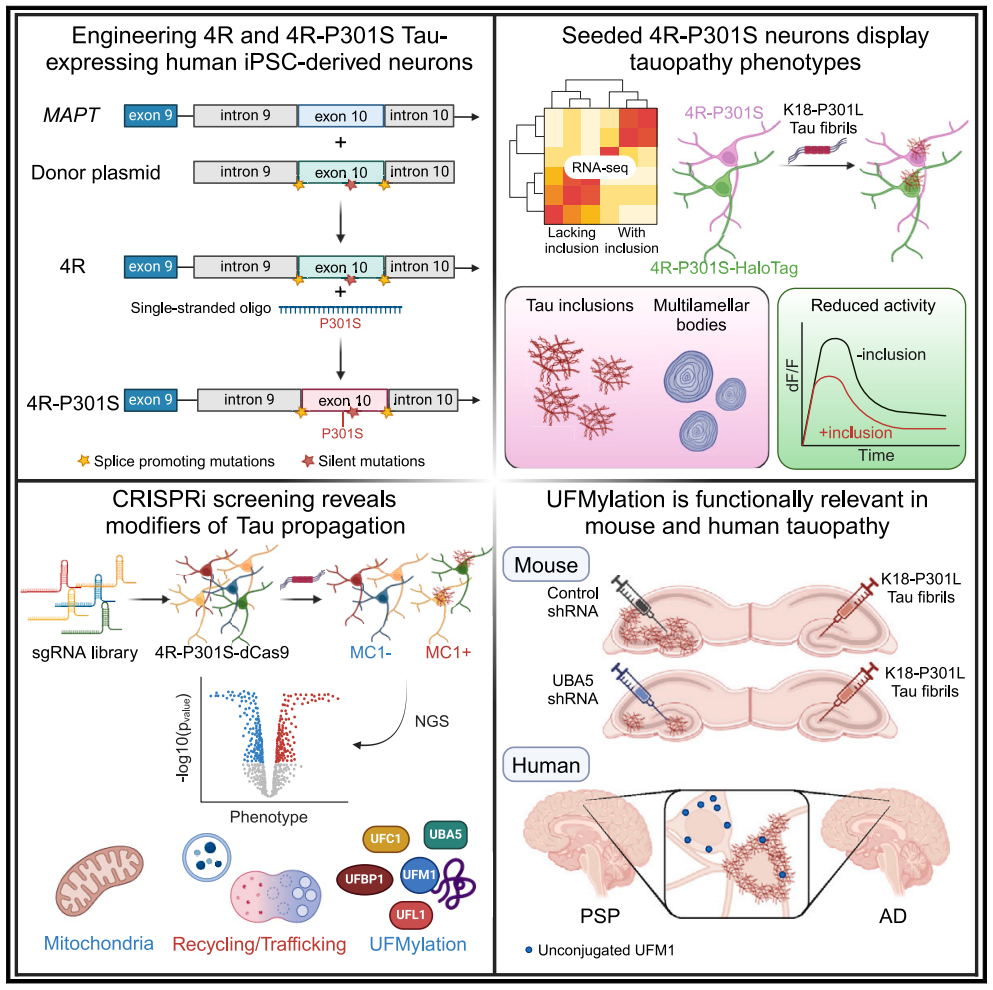
Modification of tau transmission found in human iPSC 4R segmental lesion model
In summary, the team demonstrated a robust and scalable human iPSC 4R Tau disease model. Through the engineering hiPSC cell line, 4R Tau and 4R Tau (4R-P301S) carrying P301S MAPT mutation were expressed during neuronal differentiation, and it was determined that 4R-P301S neurons could mimic the pathological characteristics of Tau disease to some extent. In addition, the research team also integrated CRISPRi and functional genome screening to identify new molecules and pathways involved in Tau disease, providing new ideas for the discovery of potential treatment strategies.
Of course, there are two main defects in this model: one is that it is unable to reproduce the characteristics of aging neurons in the human brain, and aging is one of the causes of AD; the other is that it does not reproduce the Tau lesion of AD, because the Tau carried by AD patients is 3R/4R complex, while this model only expresses 4R. These two points are also the focus of upgrading the model.
Original text link:https://doi.org/10.1016/j.cell.2024.03.015